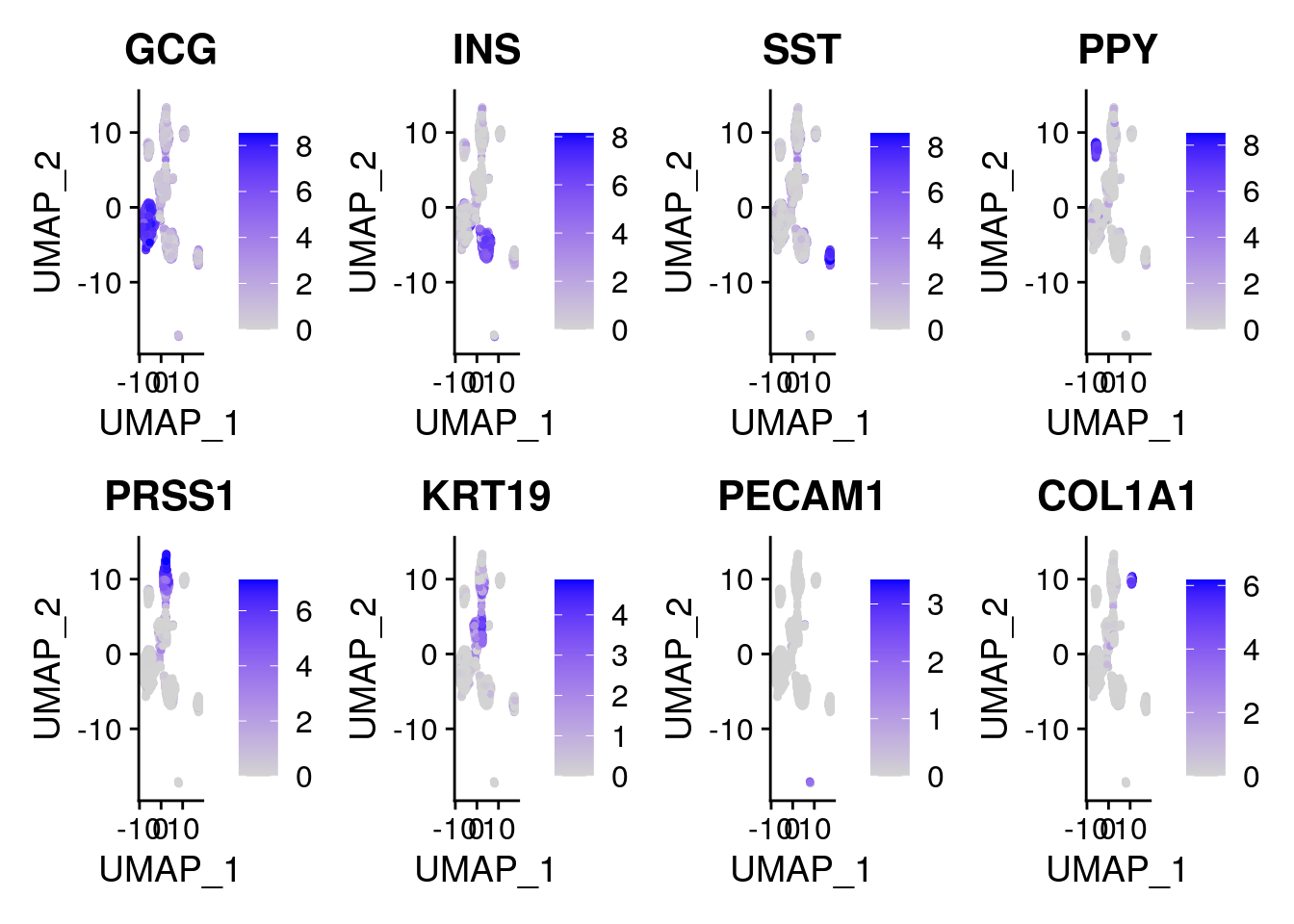
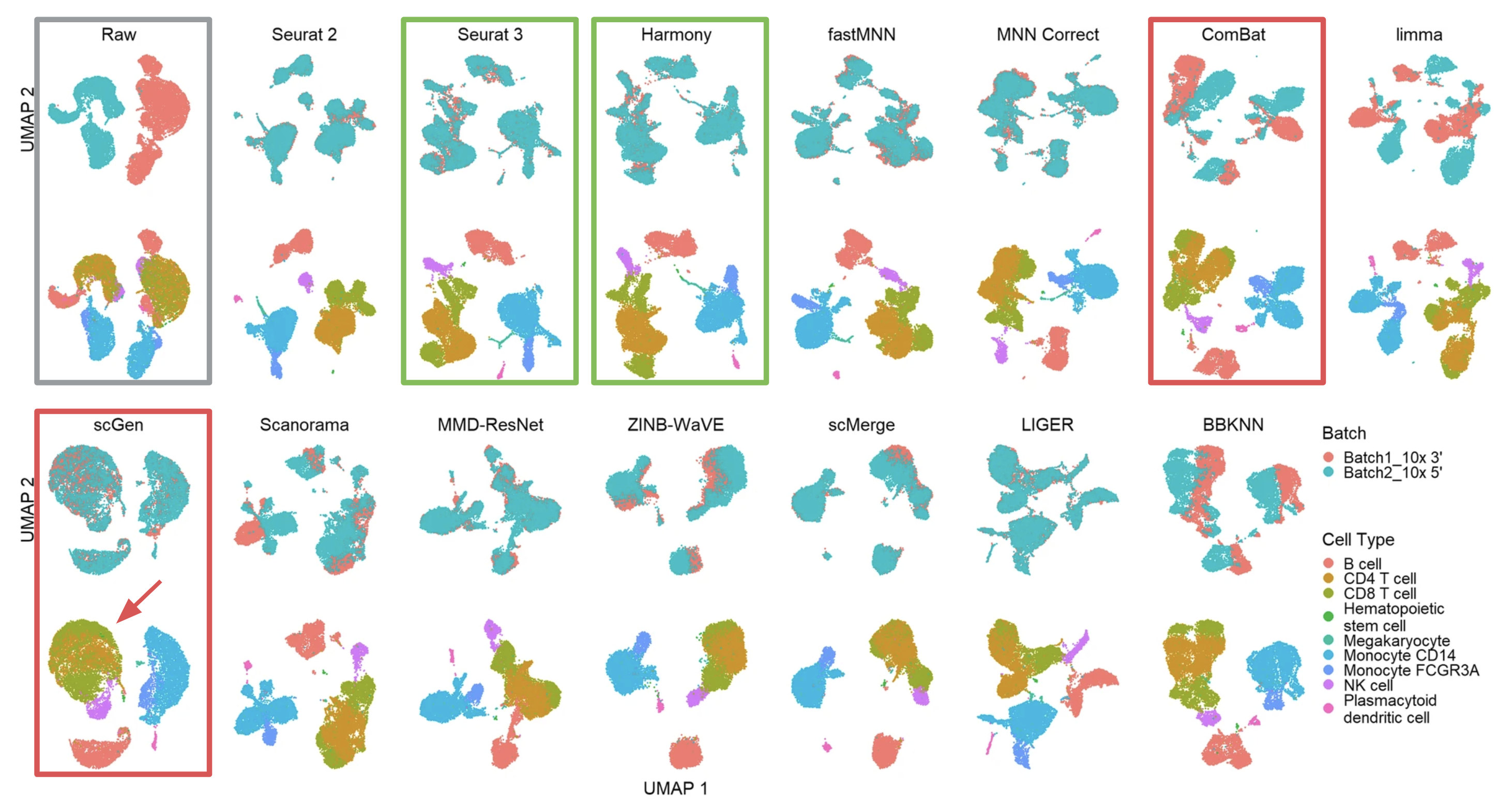
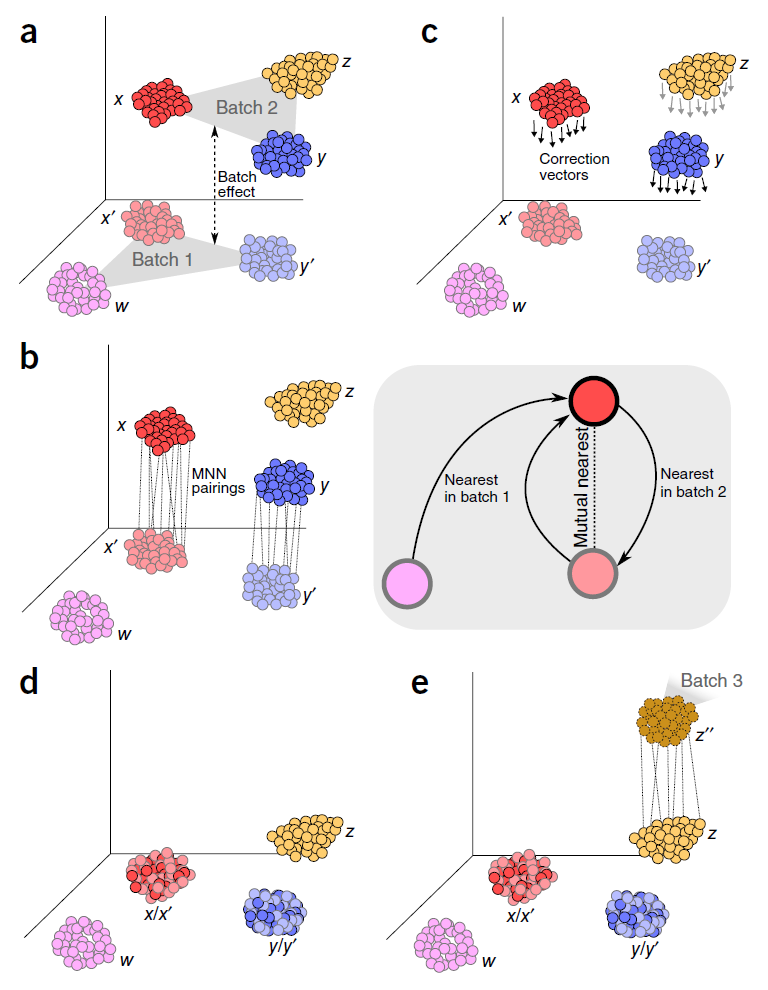
UMAP visualization of different batch effect removal methods for the... | Download Scientific Diagram

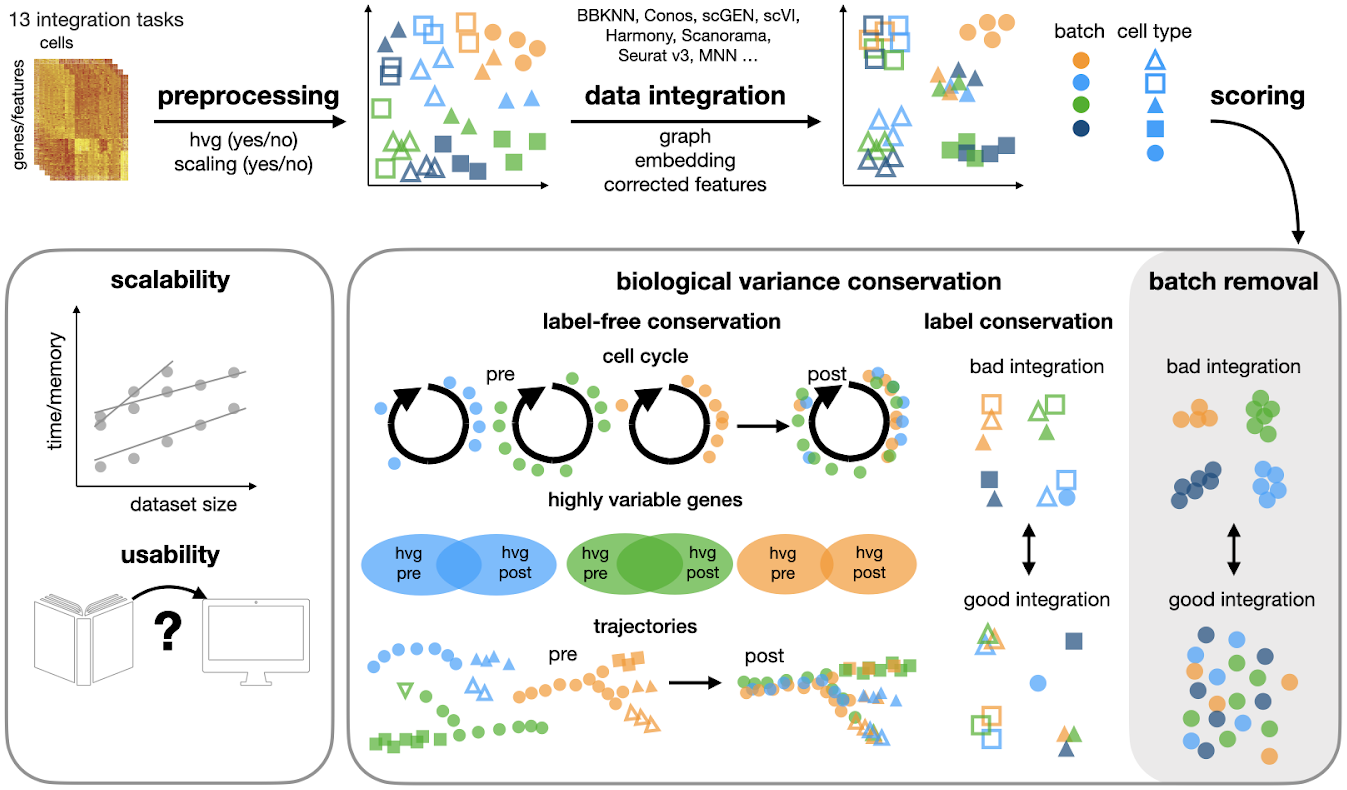
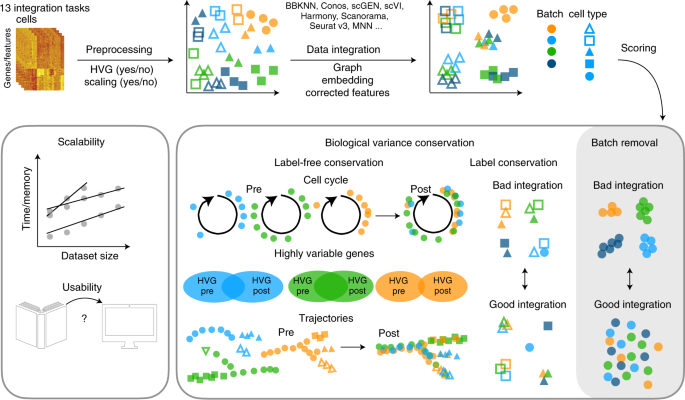
PDF) Benchmarking atlas-level data integration in single-cell genomics (2020) | Malte D Luecken | 239 Citations
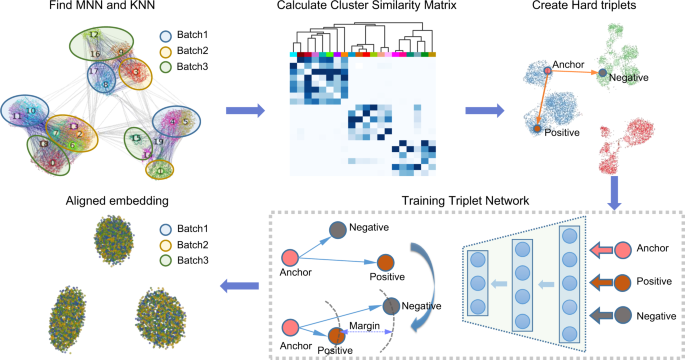
Batch alignment of single-cell transcriptomics data using deep metric learning | Nature Communications

A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity - ScienceDirect

Full article: scRAA: the development of a robust and automatic annotation procedure for single-cell RNA sequencing data
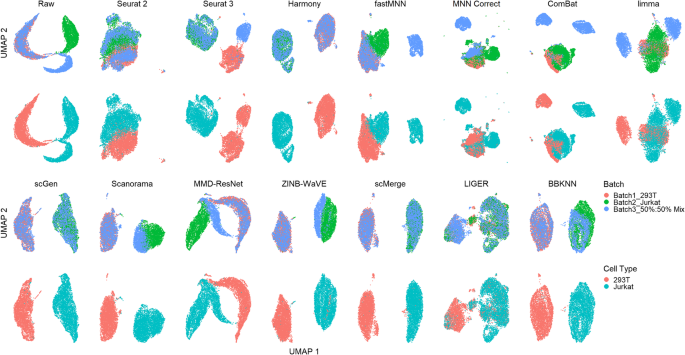
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

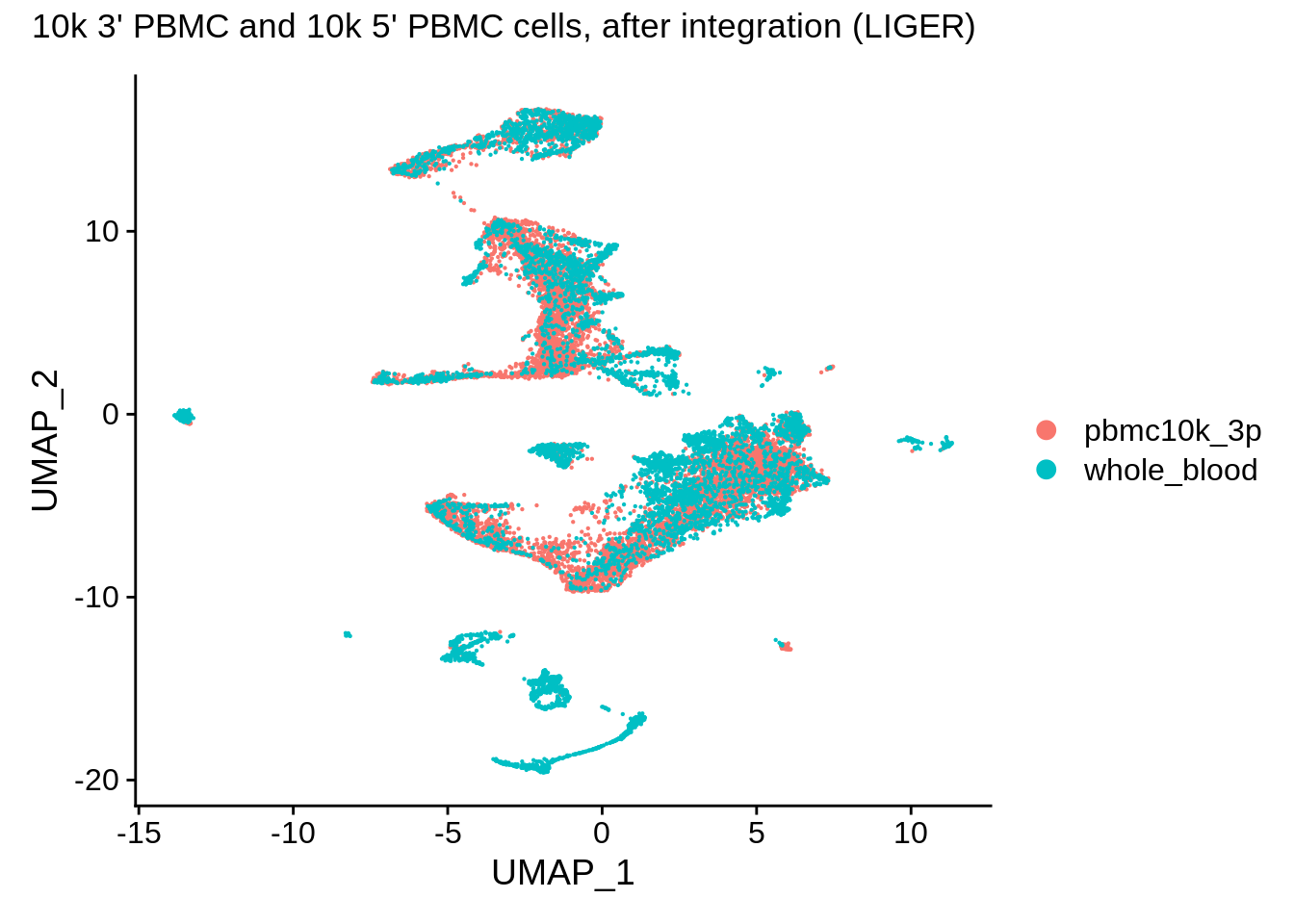
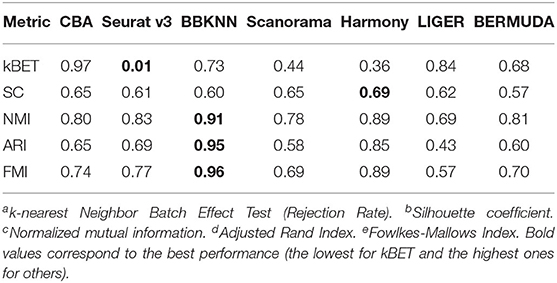
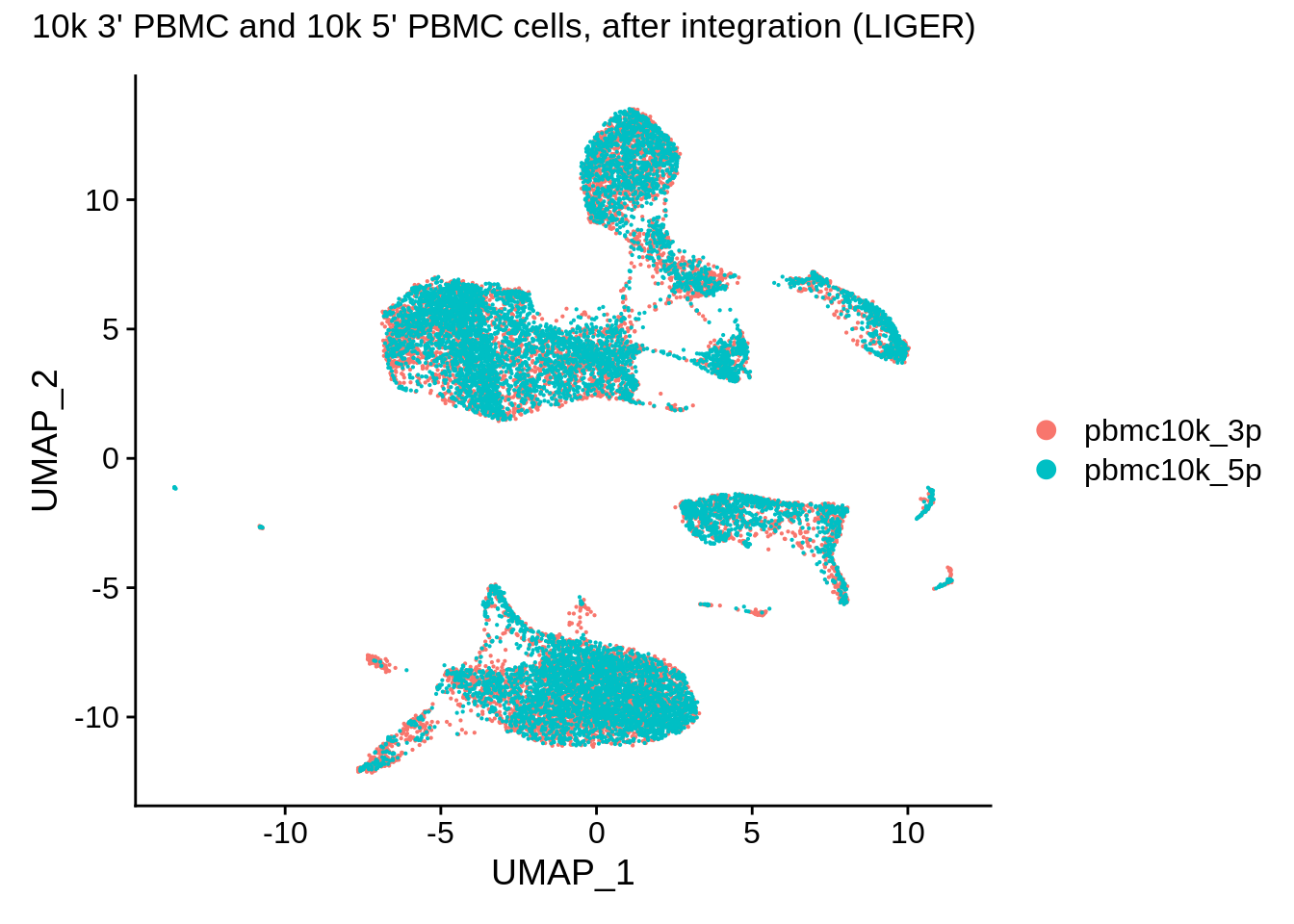
![Rethinking batch effect removing methods 系列[3] - LIGER - 知乎 Rethinking batch effect removing methods 系列[3] - LIGER - 知乎](https://pic3.zhimg.com/80/v2-f5b540c7e56f99bccd1a35127adbb4aa_1440w.webp)