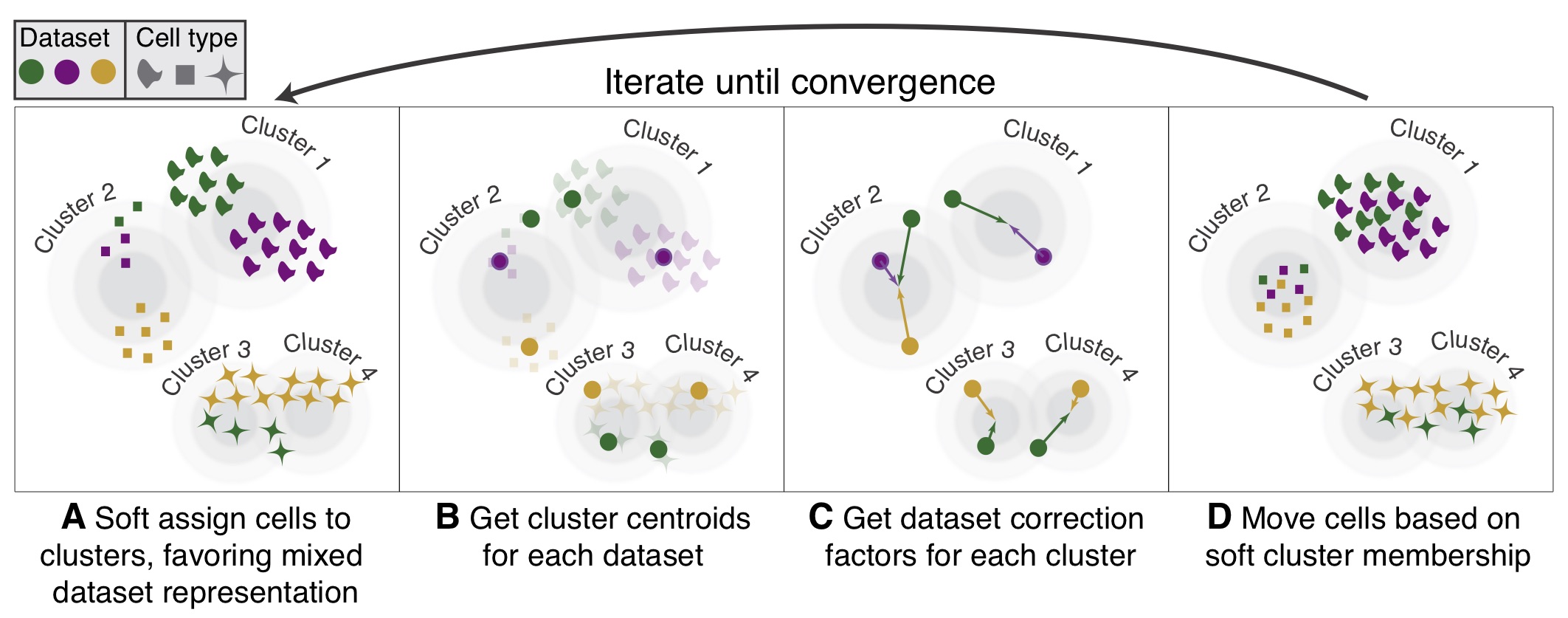
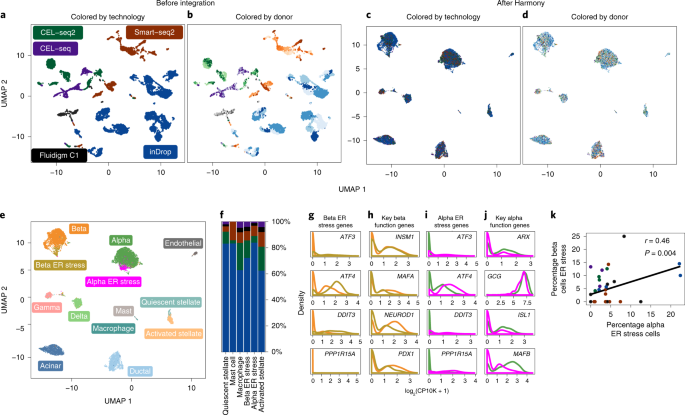
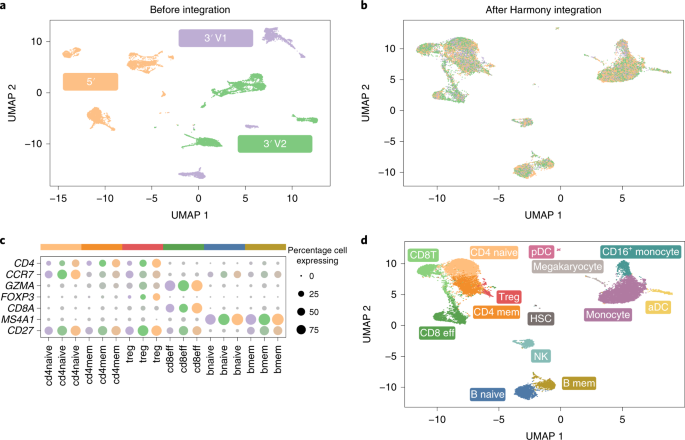
Webinar: Fast, sensitive, and accurate integration of single-cell data with Harmony | Ilya Korsunsky - YouTube

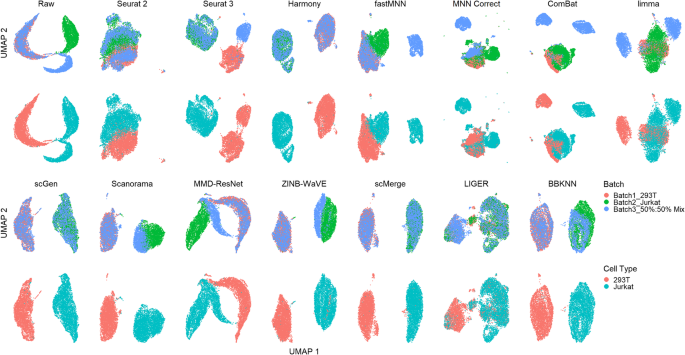
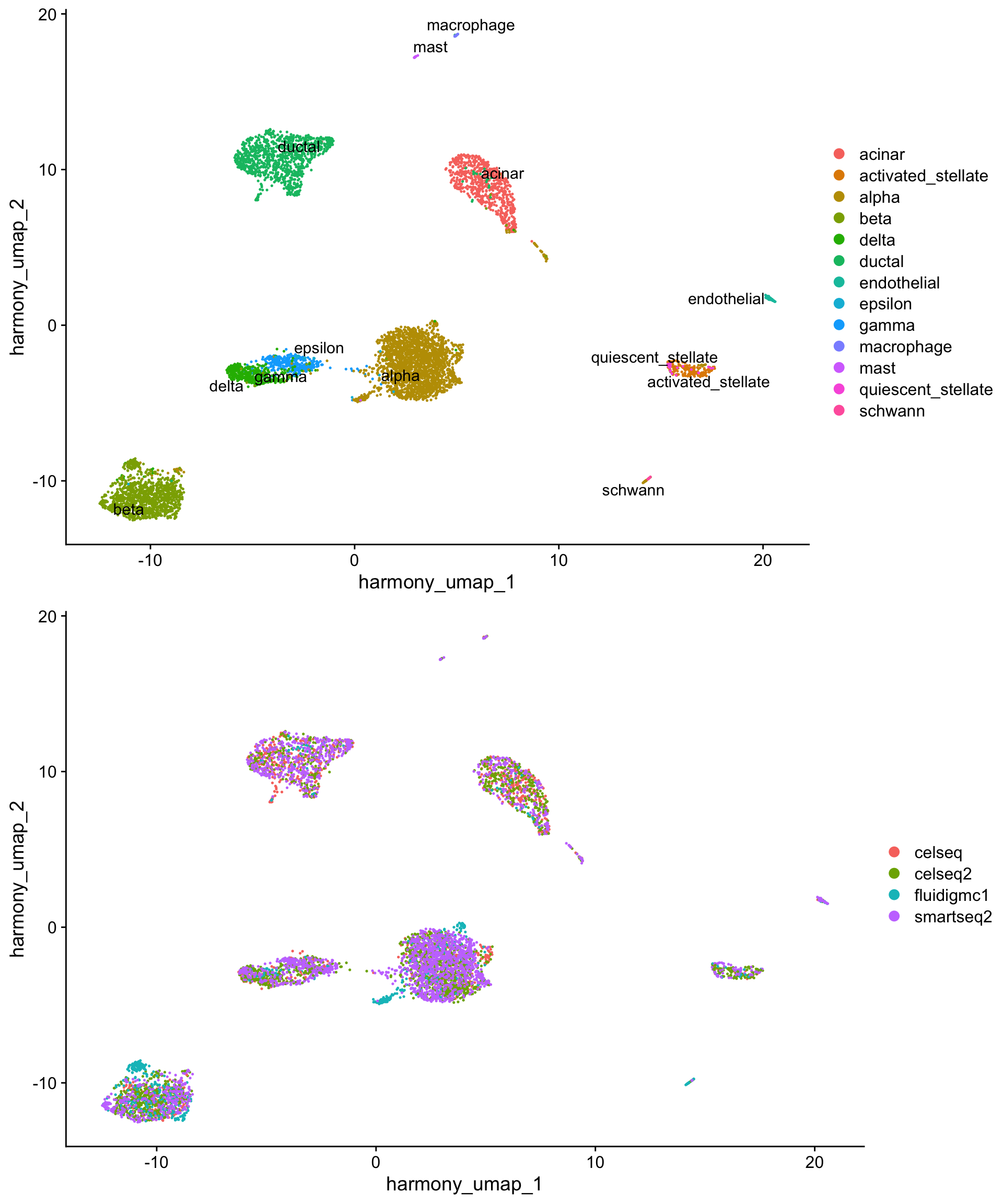
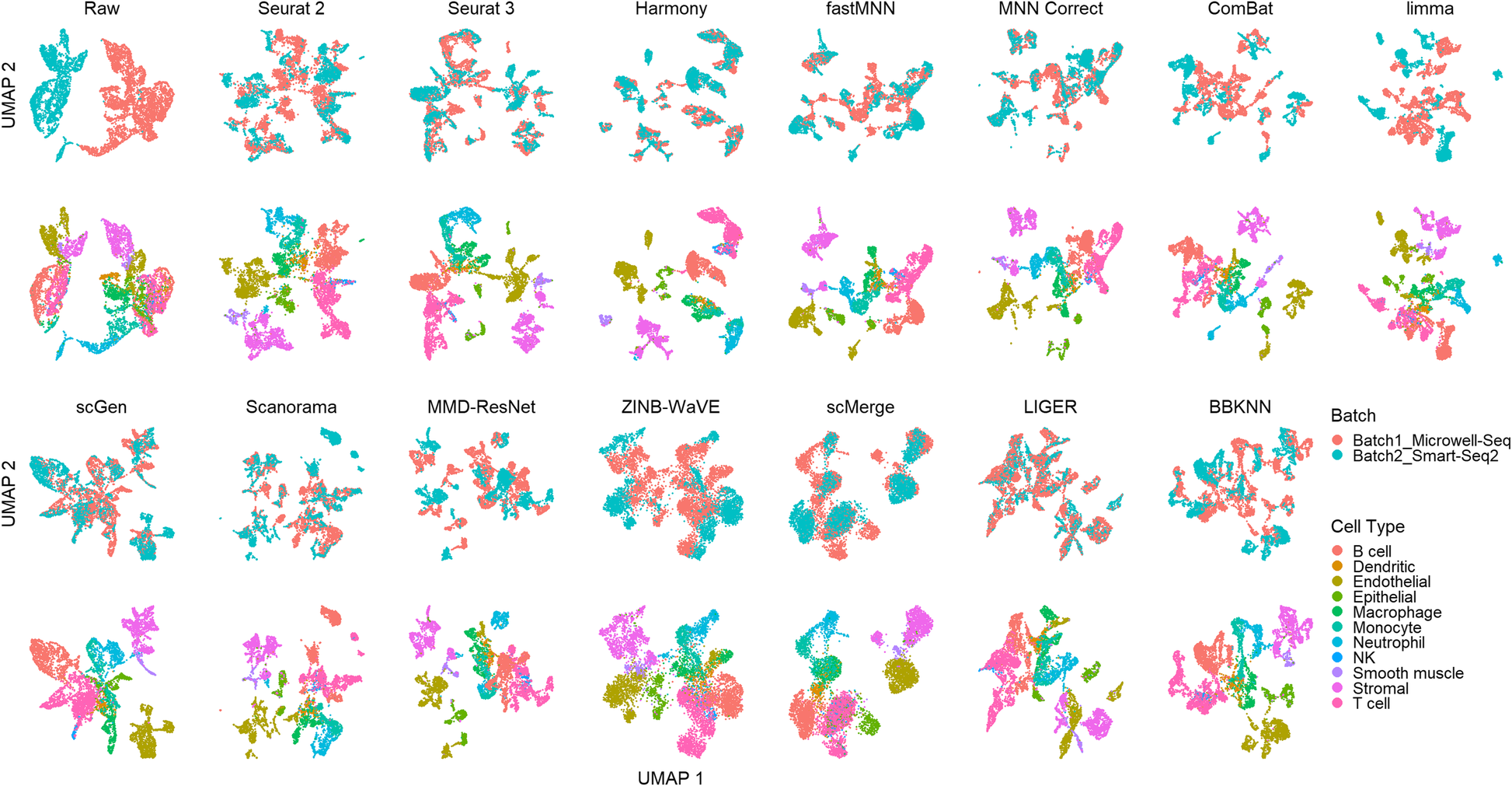
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text
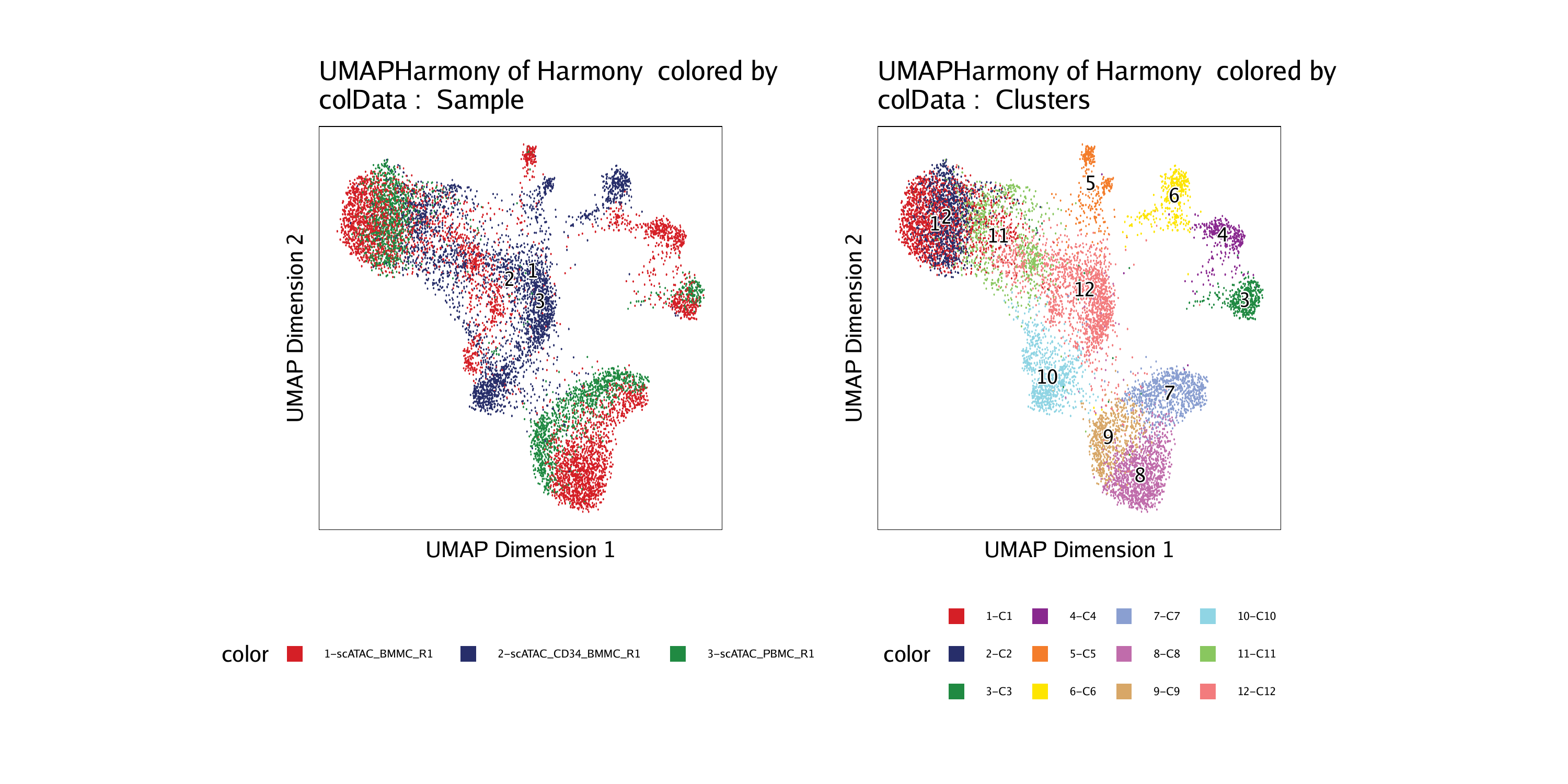
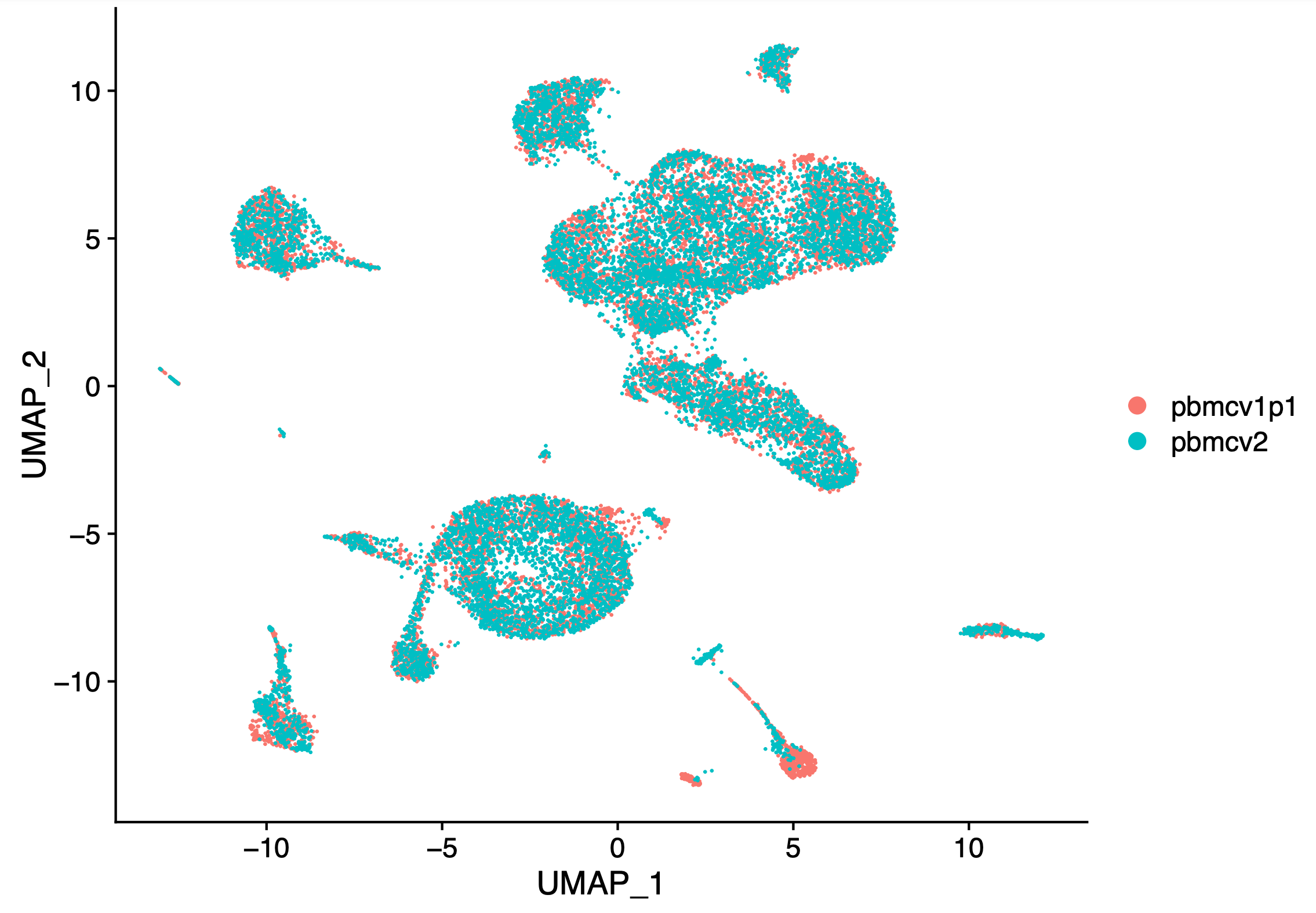
6.3 Dimensionality Reduction After Harmony | ArchR: Robust and scaleable analysis of single-cell chromatin accessibility data.
A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

A benchmark of batch-effect correction methods for single-cell RNA sequencing data | Genome Biology | Full Text

Webinar: Fast, sensitive, and accurate integration of single-cell data with Harmony | Ilya Korsunsky - YouTube
IJMS | Free Full-Text | IMGG: Integrating Multiple Single-Cell Datasets through Connected Graphs and Generative Adversarial Networks
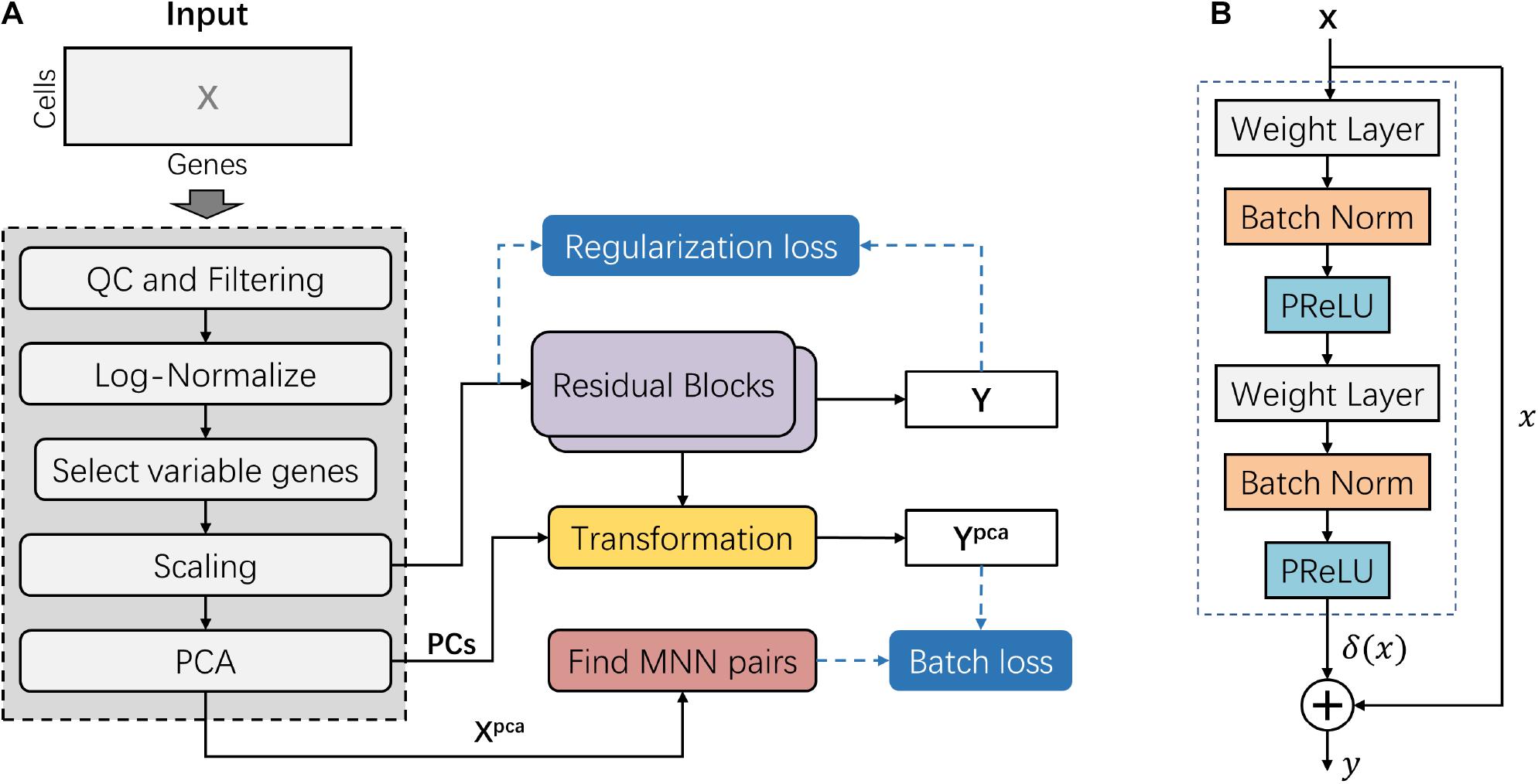
Frontiers | deepMNN: Deep Learning-Based Single-Cell RNA Sequencing Data Batch Correction Using Mutual Nearest Neighbors

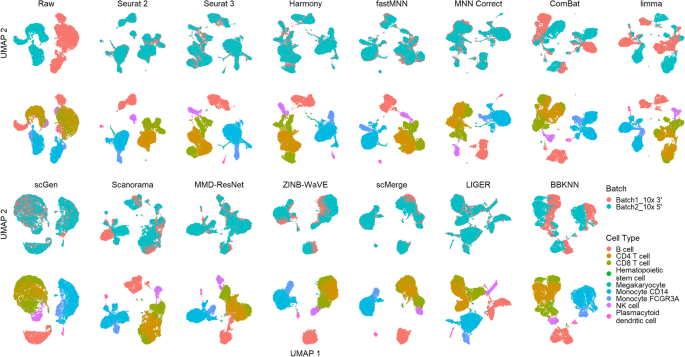
Qualitative evaluation of 14 batch-effect correction methods using UMAP... | Download Scientific Diagram

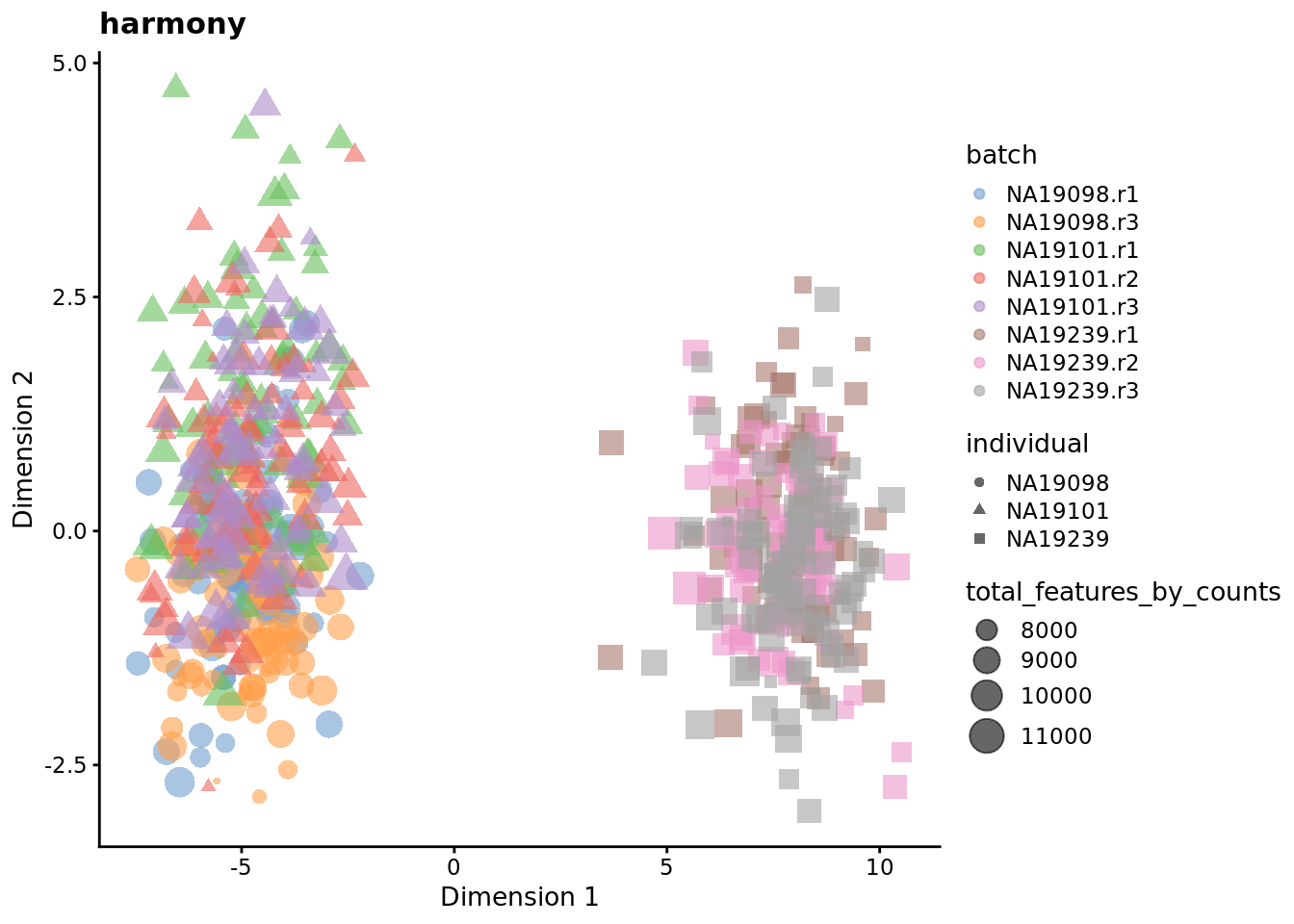
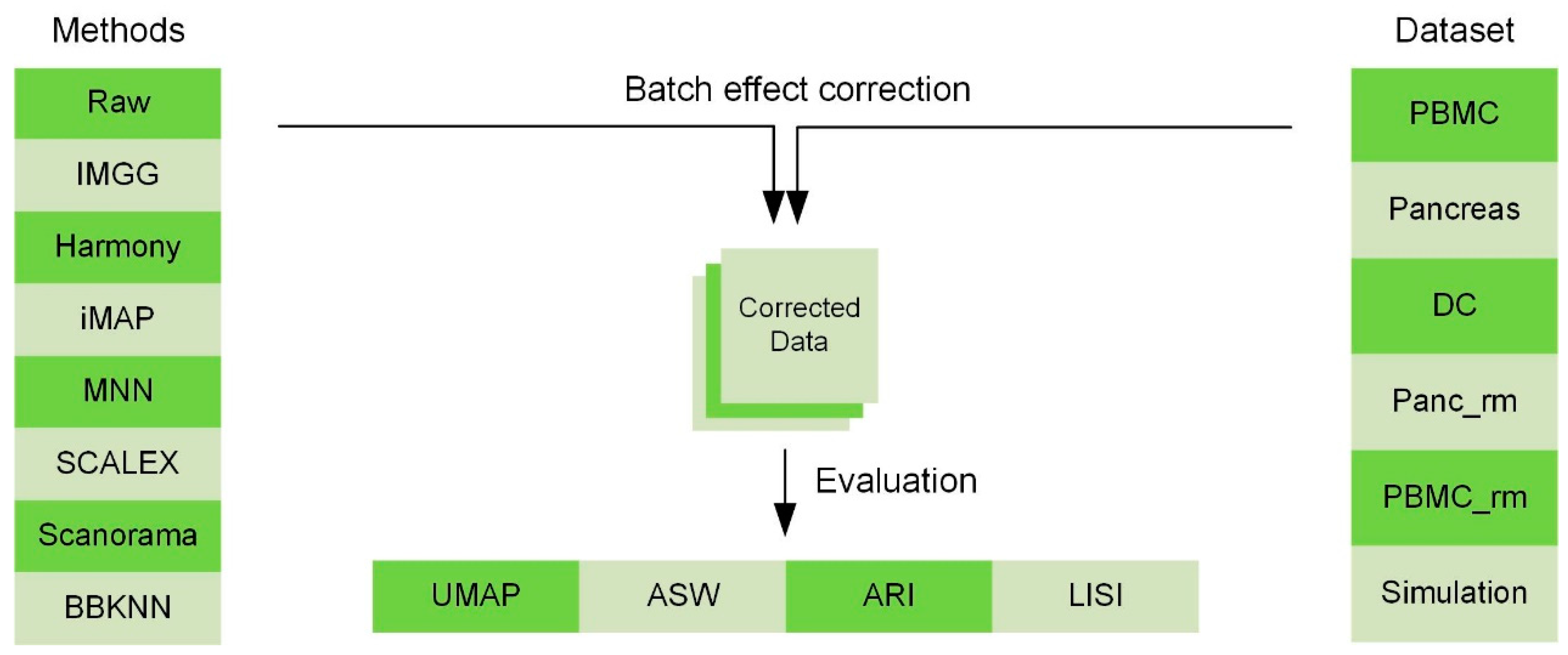
A Comprehensive Multi-Center Cross-platform Benchmarking Study of Single-cell RNA Sequencing Using Reference Samples | bioRxiv